Protein Sequence Coverage Map
Protein sequence coverage maps are visualizations used in proteomics and peptidomics to show the distribution of peptides across their parent protein. This software was developed in collaboration with Aarhus University’s Department of Food Science. Further details on the software and its use are provided below. To reference the software, please cite the accompanied review article on bioactive milk peptides.
Authors
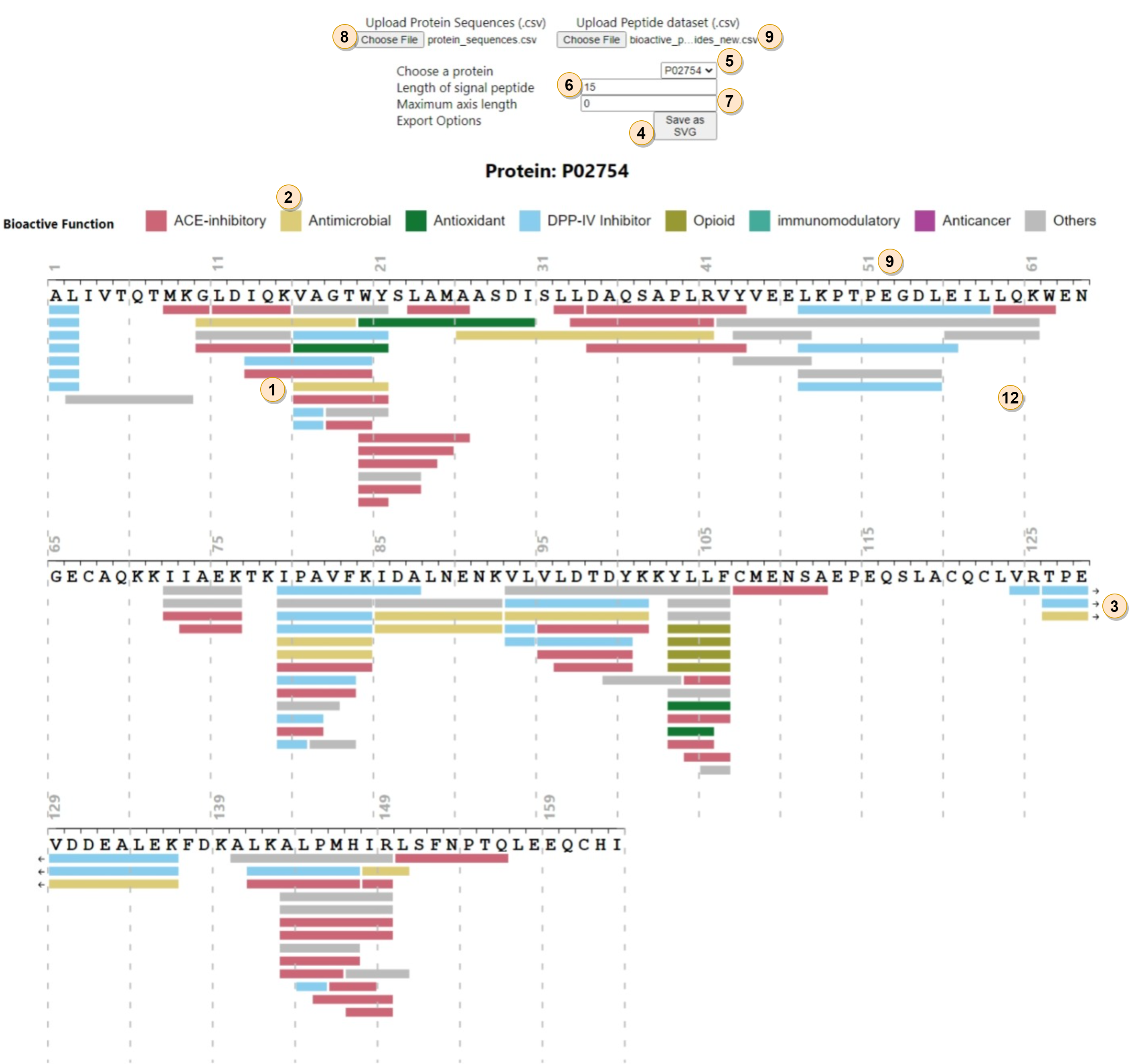
Overview
The protein sequence coverage map is widely used (1, 2, 3) to provide an overview of the peptides associated to a protein. This implementation is targeted for use cases where horizontal space is limited, e.g., in a paper.Description
The visualizations show a horizontal line (1) for each peptide, with a different color for different bioactive functions (2). The purpose of this plot is to provide an overview of all the peptides that were associated with the protein. The lines that wrap around the edges of the image are indicated with an arrow mark (3). For each amino acid "X", the longest peptide that starts with "X" is shown on top. From top to down, the length of the peptides decrease.
Currently we support exporting the plot as an SVG image (4) , which contains the plot alone without the legend or the controls.
Interaction
Choose a protein (5) : Users can choose a protein out of the list of unique values for "Entry" from the protein sequences dataset. The sequence will be built and peptides will be stacked based on this choice.
Length of signal peptide (6) : This indicates the number of amino acids to hide from the beginning of the sequence.
Max axis length (7) : This indicates the maximum number of amino acids shown per axis. If it's set to 0, then the number is calculated based on available screen width. This lets users control the width according to their horizontal space limits.
Design Choices
Color Scheme
The plot uses a categorical color scheme prescribed in Figure 4 of Qualititative Color Schemes in Paul Tol's notes. We chose this scheme as it is developed with the help of mathematical descriptions of colour differences and the two main types of colour-blind vision. The only change we've made is darkening the grey color from #DDDDDD to #BBBBBB, due to printability reasons.
As of now, the bioactive functions map to the same colors as shown in the figure, regardless of their order in any input dataset. The color codes for each bioactive function is shown in the table below.
| Bioactive Function | Color Hexcode | |
| ACE-inhibitory | #CC6677 | |
| Antimicrobial | #DDCC77 | |
| Antioxidant | #117733 | |
| DPP-IV Inhibitor | #88CCEE | |
| Opioid | #999933 | |
| immunomodulatory | #44AA99 | |
| Anticancer | #AA4499 | |
| Others | #BBBBBB |
Structure of input data
There are two datasets to be uploaded in .csv format onto fields (8) and (9), the structure of which are as follows: Protein Sequences
Should contain two columns, one for the protein ID ("Entry") and one for the protein sequence
("Sequence"). The protein ID should be unique, and should match an entry in the
UniProt database. The sequence string should have no spaces or line breaks. An example is
shown below:
| Entry | Sequence | |
| P02663 | MKFFIFTCL... | |
| P02662 | MKLLILTCL... | |
| ... | ... |
Peptides
Should contain three columns, one for the protein ID ("proteinID"), one for the peptide sequence
("Peptide") and one for the bioactive function of the peptide ("function"). The protein ID should
match one and only one entry in the protein sequences dataset.
| proteinID | peptide | function | |
| P02663 | TKVIPYVRYL | Antimicrobial | |
| P02662 | FFVAP | ACE-inhibitory | |
| ... | ... | ... |
Citation in BibTeX
To cite this article, we encourage you to use the following bibtex entry in your citation manager:
@article{doi:10.1080/10408398.2023.2240396,
author = {Søren Drud-Heydary Nielsen and Ningjian Liang and Harith Rathish and Bum Jin Kim and Jiraporn Lueangsakulthai and Jeewon Koh and Yunyao Qu and Hans-Jörg Schulz and David C. Dallas},
title = {Bioactive milk peptides: an updated comprehensive overview and database},
journal = {Critical Reviews in Food Science and Nutrition},
volume = {0},
number = {0},
pages = {1-20},
year = {2023},
publisher = {Taylor & Francis},
doi = {10.1080/10408398.2023.2240396},
note ={PMID: 37504497},
URL = {
https://doi.org/10.1080/10408398.2023.2240396
},
eprint = {
https://doi.org/10.1080/10408398.2023.2240396
}
}